Innovations in LP-EM
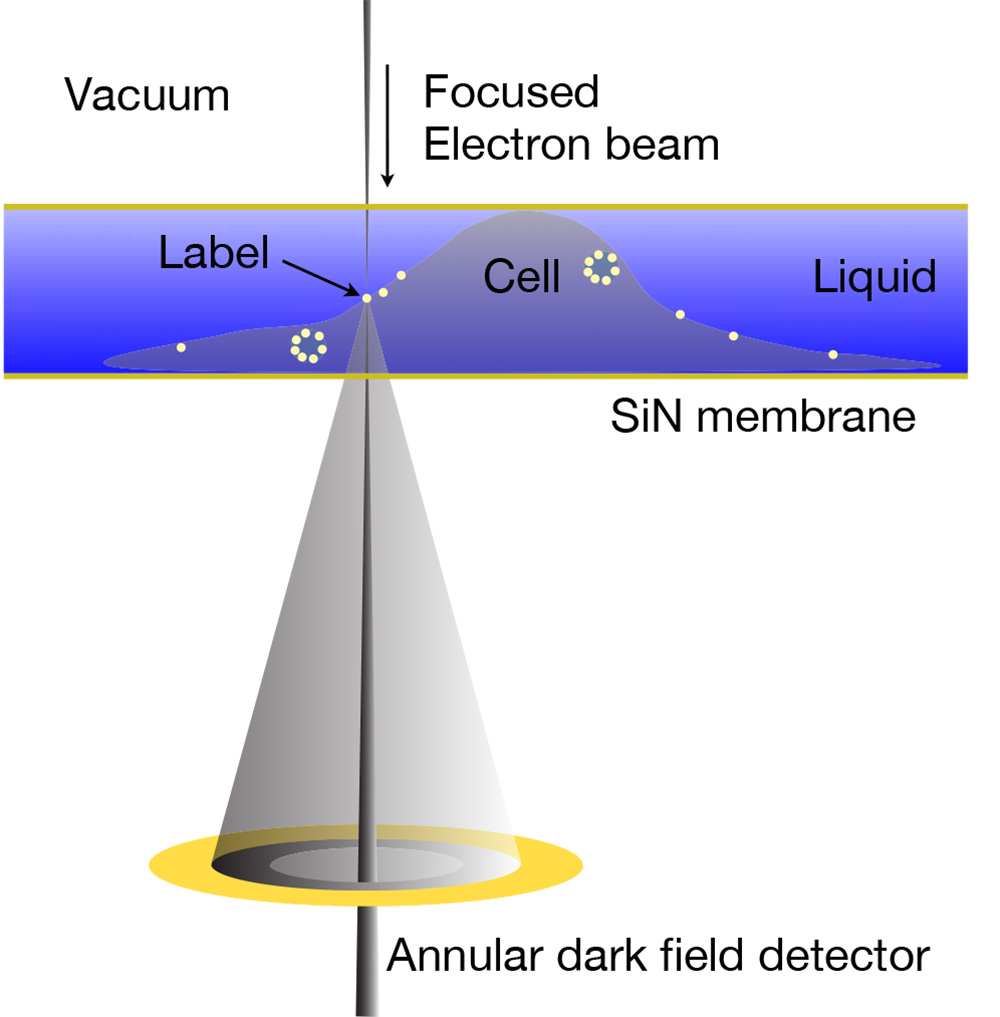
My team has developed a novel electron microscopy technology to study protein complexes in whole eukaryotic cells in their native liquid state, so-called liquid-phase electron microscopy (LP-EM). Proteins are specifically labeled with electron-dense nanoparticles such as quantum dots. The atomic number (Z) contrast of scanning transmission electron microscopy (STEM) is then applied to image the nanoparticles within a layer of liquid containing the cells. With LP-EM, we are able to distinguish labels of different sizes and compositions within the analyzed cells. LP-EM is also used to study nanomaterials in liquid, as well as protein structure.
The IEM group runs a state-of-the-art electron microscopy facility including aberration-corrected electron microscopy, environmental scanning electron microscopy, and fluorescence microscopy.
For LP-EM, we use three different approaches:
- The cells are fully enclosed in a microfluidic chamber with two SiN windows. Imaging is accomplished with STEM at 200 keV electron beam energy. See: de Jonge et al. 2009
- The sample is maintained in a saturated water vapor atmosphere, while a thin layer of water covers the cell. Environmental scanning electron microscopy (ESEM) with STEM detection is used for imaging. See: Peckys et al. 2013
- A sample in liquid is covered with a graphene sheet protecting the liquid from evaporating, and the sample is then imaged with STEM at 200 keV. See: Dahmke et al. 2017
The LP-EM approach combines much of the functionality of light microscopy with the high spatial resolution of electron microscopy. Correlative fluorescence microscopy and LP-EM are enabled using quantum dot nanoparticles as protein labels. We have extensively studied the physics of image formation of STEM in thick layers of liquid, see: Ultramicroscopy 187, 113 (2018) and Nat. Rev. Mater. 4, 61 (2019). We demonstrated that graphene enclosure of a protein sample increased the electron dose tolerance by an order of magnitude compared to cryo-electron microscopy, see: Nano Lett. 18, 7435 (2018).
Besides its application for biological samples, LP-EM has also been used to study nanomaterials in liquid to explore, for example, nanoparticle movement in thin liquid layers, self-assembly processes, and growth of nanomaterials in liquid.
A review about LP-EM is available here: Nat. Nanotechnol. 6, 695 (2011)
A review about LP-EM is available here: Nat. Nanotechnol. 6, 695 (2011), LP-EM for soft matter was reviewed here: Adv. Mater 32, 2001582 (2020).
Figure at the left: LP-EM of labeled proteins in an enclosed cell via STEM detection. From: Proc. Natl. Acad. Sci. 106, 2159 (2009).
Liquid 3D STEM
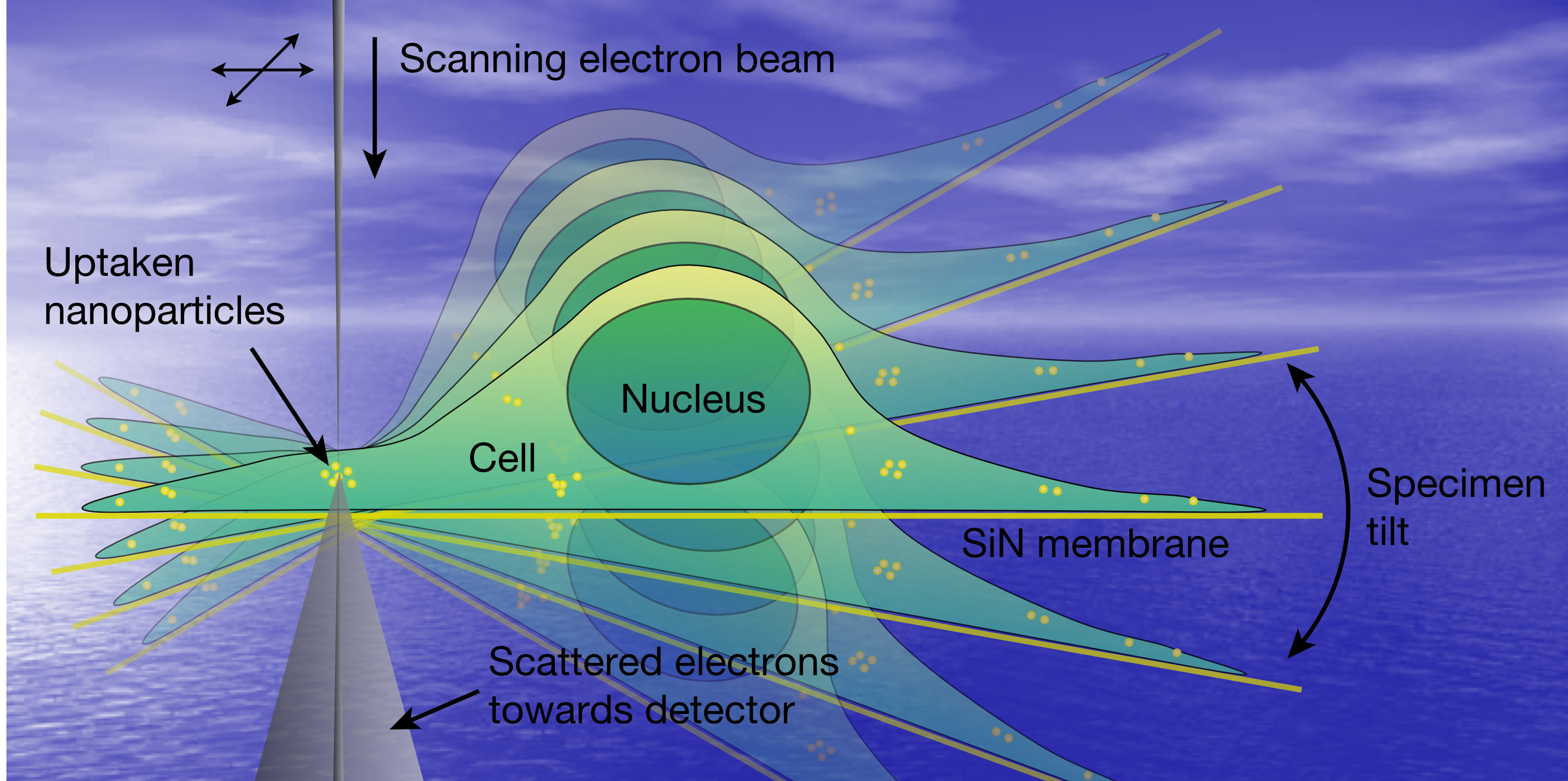
We aim to develop an electron microscopy modality capable of acquiring three-dimensional (3D) data from samples in liquid. However, standard 3D tilt-series tomography is limited on account of the high tilt-angles of up to 70°, and it is a challenge to image micrometers-thick samples, for example, whole cells. We have developed a novel 3D STEM technique for cell biology obtaining nanometer resolution on biological specimens using aberration-corrected STEM. In a manner similar to confocal light microscopy, the sample is scanned layer by layer, by changing the objective lens focus so that a focal series is recorded. Nanoscale 3D resolution results from the high beam convergence angle. We are currently improving the vertical resolution by combining focal- and tilt-series STEM. This research is conducted in conjunction with Dr. Tim Dahmen of the German Center for Artificial Intelligence and funded by the DFG in the project: “TFS-STEM: Combined tilt- and focal series for STEM tomography with a computational correction for beam blurring.” See also: Microsc. Microanal. 20, 548 (2014).
Along with Prof. Karine Masenelli-Varlot, University of Lyon, France, we further develop liquid-phase 3D STEM in a project funded by the DFG: “Liquid-phase 3D scanning transmission electron microscopy for materials science and biology”.
Figure at the left: Artist impression of 3D LP-EM. Copyright © Niels de Jonge